February 7, 2020
Updated May 11, 2020
ELECTRONIC BIOLOGY APPROACH TO FIGHT THE DEADLY WUHAN CORONAVIRUS
Fears are mounting worldwide over the cross-border spread of the new strain of coronavirus (denoted as 2019-nCoV) originated in Wuhan, the largest city in central China. This deadly virus currently spread in 28 countries, with more than 24,500 cases of which more than 3000 are in critical conditions and at least 493 deaths. The genome sequencing of the new coronavirus, published in record time in mid-January, has given way to a race to gain protection from an epidemic whose scope is still unknown. The most urgent questions surrounding the outbreak are (i) what is the origin and/or natural reservoir of the virus? (ii) is it easily transmitted from human to human? and (iii) what are the potential diagnostic, therapeutic and vaccine targets?
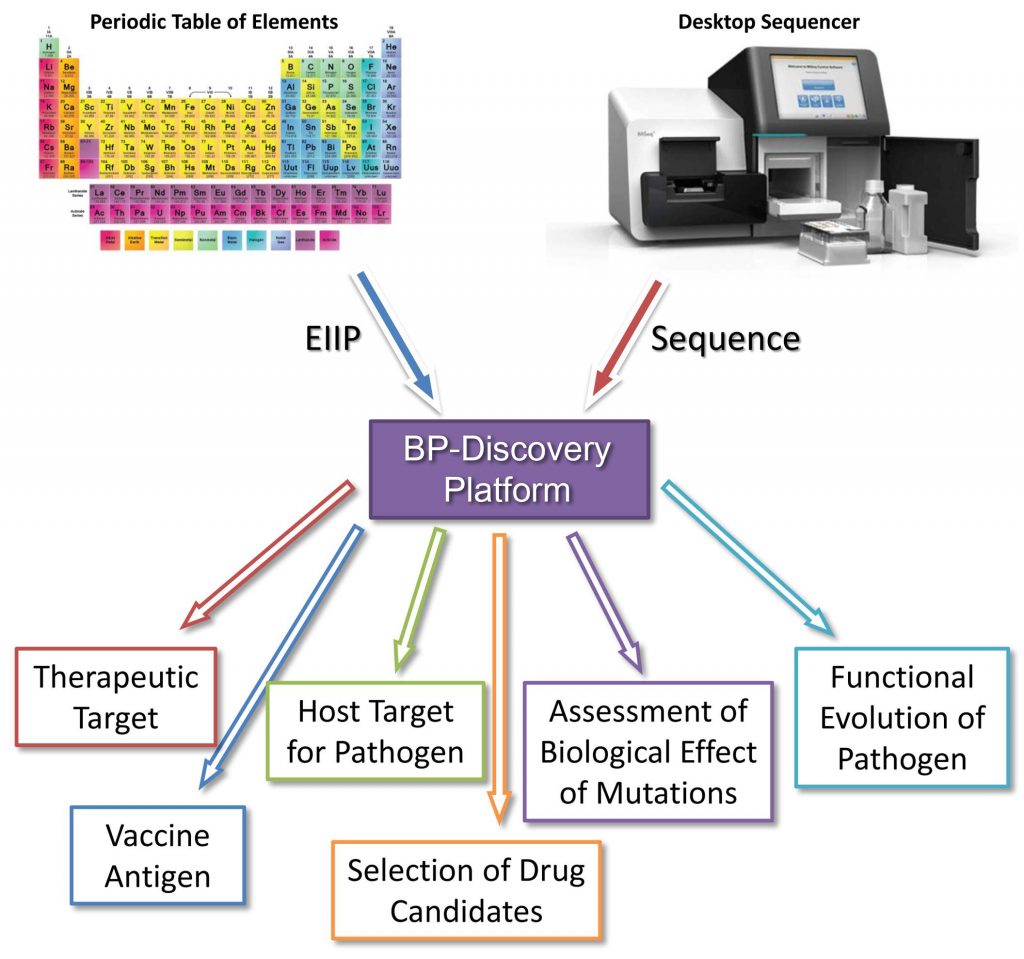
Researchers at Biomed Protection Ltd (Galveston, USA), the Galveston National Laboratory and the University of Texas Medical Center (Galveston, USA) and the Research Center for Animal Health (Barcelona, Spain), tried to answer some of these important questions in the recently published article “Use of the informational spectrum methodology for rapid biological analysis of the novel coronavirus 2019-nCoV: prediction of potential receptor, natural reservoir, tropism and therapeutic/vaccine target” (https://f1000research.com/articles/9-52). In contrast to other initial studies of Wuhan coronavirus, which mainly are based on the simple comparisons of sequences from this virus and other known coronaviruses, in this article analysis was performed using the novel Electronic Biology approach allowing study of the essential physico-chemical properties of viral proteins.
Results of this first published molecular analysis of the Wuhan coronavirus suggest the following: (i) the newly emerging 2019-nCoV is highly related to SARS-CoV and, to a lesser degree, MERS-CoV; (ii) civets and poultry are potential candidates for the natural reservoir of the 2019-nCoV and could be use as the animal models for further analysis of this virus, (iii) human actin proteins possibly participate in infection and pathogenesis of 2019-nCoV allowing direct cell-to-cell transmission of the virus, and (iv) drugs which interact with actin proteins (e.g. ibuprofen) should be investigated as possible therapeutics for treatment of 2019-nCoV infection. The analysis also revealed domain of the viral S protein which could be essential for human infection. This represents the potential Achilles Heel of the Wuhan coronavirus which could be targeted with the vaccine. These results obtained in silico will be further experimentally tested by US and Spanish virologists.
The Electronic Biology platform (e-Bio) is independent of the “wet-lab” experimentation and can be applied to multiple disease area. The e-Bio platform allows discovery of the diagnostic, therapeutic and vaccine targets for novel emerging pathogens immediately after their isolation and sequencing.
25 Feb, 2020
SHORT REPORT, Virology
COVID-19 Orf3b protein: the putative biological function and the therapeutic target
https://www.researchsquare.com/article/rs-15042/v1

05.05.2020
ChemRxiv
Drug Repurposing for Candidate SARS-CoV-2 Main Protease Inhibitors by a Novel in Silico Method
https://doi.org/10.26434/chemrxiv.12248561.v1

14.05.2020
F1000 Research
Prediction of the effectiveness of COVID-19 vaccine candidates
https://f1000research.com/articles/9-365/v1

15.08.2020
Journal of Proteome Research
The Biological Rationale for the Repurposing of BCG Vaccine against SARS-CoV-2
https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00410

DISCOVERY OF THE SARS-C0V-2 RECEPTOR
Veljkovic, V.; Vergara-Alert, J.; Segalés, J.; Paessler, S. Use of the informational spectrum methodology for rapid biological analysis of the novel coronavirus 2019-CoV: prediction of potential receptor, natural reservoir, tropism and therapeutic/vaccine target. F1000Research 2020; 9:52-65. https://doi.org/10.12688/f1000research.22149.1
Received 20 January 2020
Accepted 25 January 2020
Accepted manuscript posted online 27 January 2020
Abstract
The performed in silico analysis indicates that the newly emerging SARS-CoV-2 is closely related to severe acute respiratory syndrome (SARS)-CoV and, to a lesser degree, Middle East respiratory syndrome (MERS)-CoV. Moreover, the well-known SARS-CoV receptor (ACE2) might be a putative receptor for the novel virus.
Downloads: 1,117
Citation: 30
Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. https://doi .org/10.1128/JVI.00127-20.
Received 22 January 2020
Accepted 28 January 2020
Accepted manuscript posted online 29 January 2020
Abstract
Here, we analyzed the potential receptor usage by 2019-nCoV, based on the rich knowledge about SARS-CoV and the newly released sequence of 2019-nCoV. First, the sequence of 2019-nCoV RBD, including its receptor-binding motif (RBM) that directly contacts ACE2, is similar to that of SARS-CoV, strongly suggesting that 2019-nCoV uses ACE2 as its receptor.
Downloads: 268,686
Citation: 3,590
Media Contact: veljko@biomedprotection.com

